How scientists are creating synthetic life from scratch
By Susannah Locke
Over the past decade, the ease of sequencing and creating DNA has improved so much that the possibilities of genetic engineering have expanded tremendously.
Researchers can now go way beyond the slight tinkering they've been done in the past — like adding or deactivating a single gene. Instead, some scientists are now focusing on broadly creating and re-engineering living things wholesale to improve our environment, our energy, and our health
Welcome to the strange new world of synthetic biology, in which living things are a tool to be manipulated for practical ends. It's a world in which, someday, organisms designed from scratch could convert waste into fuel or enter people's bodies to kill cancer.
Some scientists see synthetic biology as the best bet to tackle some of the world's most pressing problems — like the ever-increasing demand for food and energy. But the prospect of possible mishaps, not to mention concerns about tinkering with life to begin with, are certainly there, too. Here's a primer on synthetic biology.
What is synthetic biology, anyway?
The term "synthetic biology" generally refers to the engineering of new biological tools for practical purposes. If that sounds a lot like the existing practice of genetic engineering — well, that makes sense, because it is.
Many scientists simply refer to synthetic biology as "genetic engineering on steroids" (to quote Jim Collins, a pioneer in the field). But there's not always a clear line at which ho-hum genetic engineering flips over into synthetic biology territory.
In general, if you're genetically engineering foods like corn and soy by adding or modifying a single gene, that's not synthetic biology. But if you're adding in a whole suite of genes or creating an entirely new genetic code that doesn't exist anywhere in nature, then you're definitely entering synthetic biology territory.
Synthetic biologists use a variety of approaches, some of which can overlap:
1) Removing inefficiencies in cells
Some researchers are trying to remove inefficiencies from cells that are a byproduct of the haphazard nature of evolution. For example, if you're engineering bacteria to produce biofuels, you want the process to be as efficient as possible. Researchers also use this kind of approach to find the limits of life — how simple or how different can something be and still be alive?
Some researchers are trying to remove inefficiencies from cells that are a byproduct of the haphazard nature of evolution. For example, if you're engineering bacteria to produce biofuels, you want the process to be as efficient as possible. Researchers also use this kind of approach to find the limits of life — how simple or how different can something be and still be alive?
2) Combining genetic sequences in extreme ways
Some researchers want to combine many genes from various organisms to make new tools. For example, some who are interested in having algae make fuel think that combining DNA across many algae strains will be the key that has eluded them so far.
Some researchers want to combine many genes from various organisms to make new tools. For example, some who are interested in having algae make fuel think that combining DNA across many algae strains will be the key that has eluded them so far.
3) Designing new "living machines"
Others are trying to design living machines by reprogramming DNA into logical circuits to make them function like miniature computers. For example, researchers have gotten cells to do arithmetic and show their answers by lighting up in a certain color.
Others are trying to design living machines by reprogramming DNA into logical circuits to make them function like miniature computers. For example, researchers have gotten cells to do arithmetic and show their answers by lighting up in a certain color.
What could we do with synthetic biology?

Green fuels is one area where synthetic biology could have a major impact. AFP/Getty Images
Like any new technology, it's difficult to tell exactly where synthetic biology will have its biggest impact. But there are a few big areas of interest, which right now are in medicine, energy, food, and environmental remediation.
1) Medicine
Synthetic biology might one day let scientists program cells to precisely detect and kill cancer cells. Or perhaps program cells to self-assemble into spare organs for transplants. Some scientists are already using partially synthetic organisms to manufacture drugs that are otherwise impractical to make.
Synthetic biology might one day let scientists program cells to precisely detect and kill cancer cells. Or perhaps program cells to self-assemble into spare organs for transplants. Some scientists are already using partially synthetic organisms to manufacture drugs that are otherwise impractical to make.
2) Food and fragrances
In theory, new techniques could allow researchers to design yeast to make the perfect beer or wine. Or create food in the lab more efficiently than growing it on land. "We can design better and healthier proteins than we get from nature," biologist and entrepreneur Craig Venter told the New York Times.
In theory, new techniques could allow researchers to design yeast to make the perfect beer or wine. Or create food in the lab more efficiently than growing it on land. "We can design better and healthier proteins than we get from nature," biologist and entrepreneur Craig Venter told the New York Times.
Already, synthetic biology companies are selling orange and grapefruit flavorings produced by yeast. And the company Evolva makes yeast-generated artificial vanilla flavoring and is working on better tasting sugar substitutes.
3) Energy and environment
Another possibility is that synthetic biologists could program cells to produce usable fuel. For example, Exxon Mobile has a partnership with Synthetic Genomics (co-founded by Craig Venter) to research fuel from algae. Ideally, helpful organisms would eat things we don't need, like non-edible plant matter. Even more ideally, they'd eat the extra carbon dioxide in the atmosphere that's warming the planet. Or toxic waste or oil from oil spills.
Another possibility is that synthetic biologists could program cells to produce usable fuel. For example, Exxon Mobile has a partnership with Synthetic Genomics (co-founded by Craig Venter) to research fuel from algae. Ideally, helpful organisms would eat things we don't need, like non-edible plant matter. Even more ideally, they'd eat the extra carbon dioxide in the atmosphere that's warming the planet. Or toxic waste or oil from oil spills.
4) The weird stuff
How about some microbes that live on your skin to prevent you from getting oily and smelly? How about some other ones that secrete the perfume of your choice? How about some that quickly break down cholesterol so it won't clog people's arteries?
so it won't clog people's arteries?
How about some microbes that live on your skin to prevent you from getting oily and smelly? How about some other ones that secrete the perfume of your choice? How about some that quickly break down cholesterol
 so it won't clog people's arteries?
so it won't clog people's arteries?What have scientists done with synthetic biology so far?
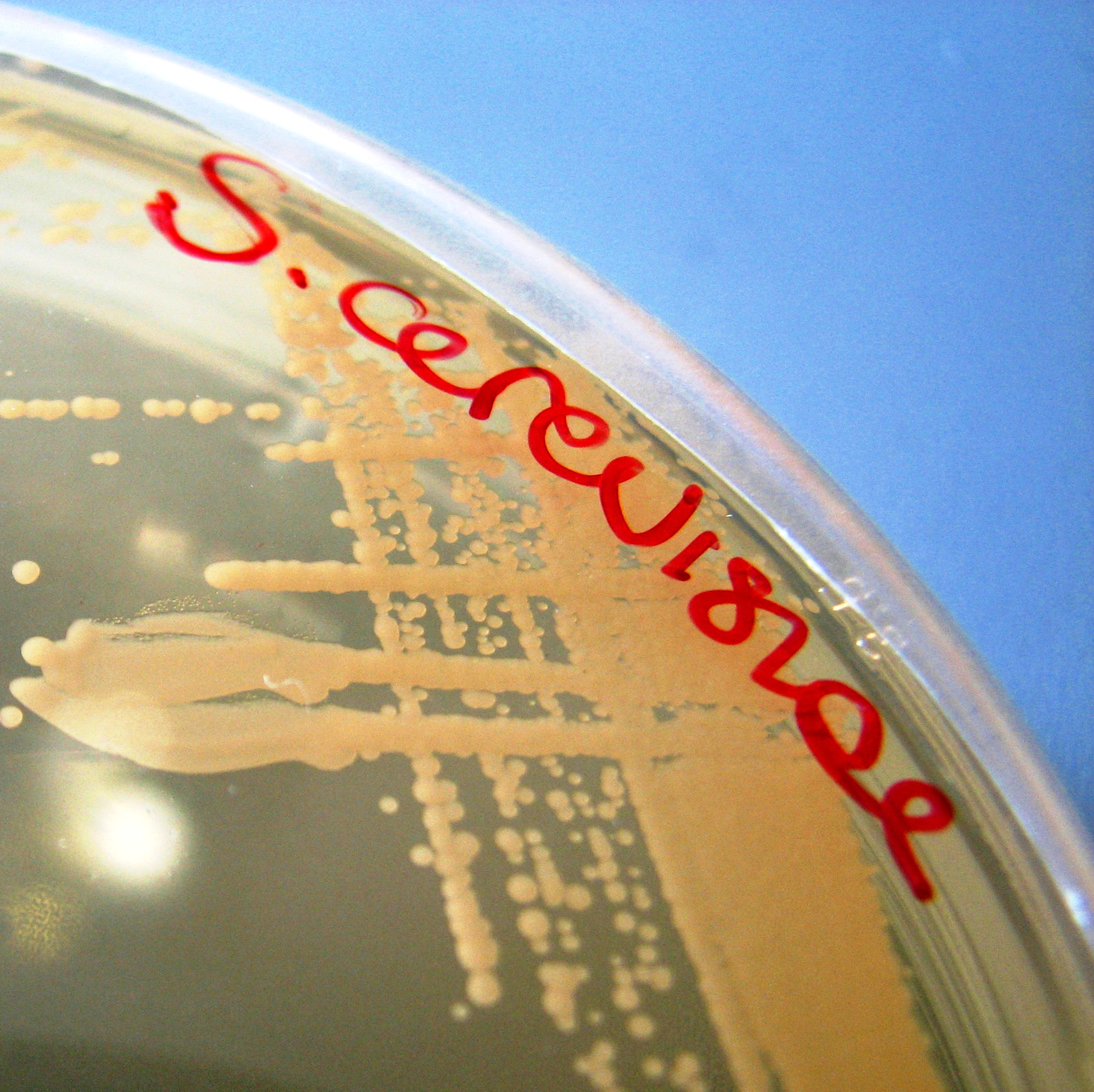
Yeast growing on a petri dish. Yeast is a major player in synthetic biology these days. Rising Damp/Flickr
We're not yet at the point of designer cells that kill cancer or turn plastic waste into fuel. But scientistshave done a few exciting things already:
1) A more efficient process for making anti-malarial drugs
Artemisinin is one of the most effective drugs to treat malaria. But for a long time, it had to be derived from the sweet wormwood plant Artemisia annua — a slow and expensive process.
Artemisinin is one of the most effective drugs to treat malaria. But for a long time, it had to be derived from the sweet wormwood plant Artemisia annua — a slow and expensive process.
That changed in 2013, when pharmaceutical firm Sanofi used synthetic biology to produce artemisinin at an industrial scale. The company did this by taking the plant's genes for making artemisinic acid and putting them in yeast, allowing them to produce the drug more quickly and efficiently. The effort is widely cited as the first large-scale drug project to use synthetic biology and as a major achievement for the field.
2) Creating bacteria from scratch
In 2010, researchers at the J. Craig Venter Institute published the results of a 15-year, $40 million project to make the first synthetic cell. How did they do it? They took the genomic code from one bacterial species, made it in a lab from scratch, and then put it into an entirely different species — that lived.
In 2010, researchers at the J. Craig Venter Institute published the results of a 15-year, $40 million project to make the first synthetic cell. How did they do it? They took the genomic code from one bacterial species, made it in a lab from scratch, and then put it into an entirely different species — that lived.
The genome they made also included some deleted genes and new sequences that acted as watermarks. And, all in all, the scientists created the first life-form living on completely synthetic genetic material. They called it the first synthetic cell. (However, they didn't say that they created life itself from scratch. Had they put the DNA into an already-dead cell, nothing would have happened.)
This work didn't create bacteria that was useful for any particular purpose, but it was an important proof of principle that a cell can survive on lab-made DNA.
3) Creating yeast from scratch
In 2014, a team of researchers from many institutions including Johns Hopkins University revealed that they had synthesized an entire yeast chromosome from scratch. And the chromosome functioned when put back into a yeast cell. This was an especially impressive feat because yeast's genetic material is more complex than bacteria's.
In 2014, a team of researchers from many institutions including Johns Hopkins University revealed that they had synthesized an entire yeast chromosome from scratch. And the chromosome functioned when put back into a yeast cell. This was an especially impressive feat because yeast's genetic material is more complex than bacteria's.
The scientists called the DNA they made a "designer chromosome" because they deleted any sequences that they found unnecessary and added in elements that will allow future researchers to easily delete any gene they want. The goal is to rewrite the entire yeast genome in five years. So far, they've done about 3 percent of it, by length. Only 15 more chromosomes to go.
So how do you design an organism from scratch?
The main tool here is the computer. Researchers work with the code of existing organisms' genetic material as essentially a text file, tweaking it, deleting parts, adding parts, adding parts from other organisms, whatever they want.
Then they need to take that information and turn it into physical DNA. So they use a DNA synthesis machine that creates actual DNA from the necessary molecules. DNA that has been made by a machine is considered "synthetic DNA."
The researchers have to get that DNA into the organism of choice, and the techniques here can vary depending on the type of cell. Shorter chunks of DNA are easier to work with than longer chunks, which is why you see many small DNA pieces in the graphic below (from Nature News).
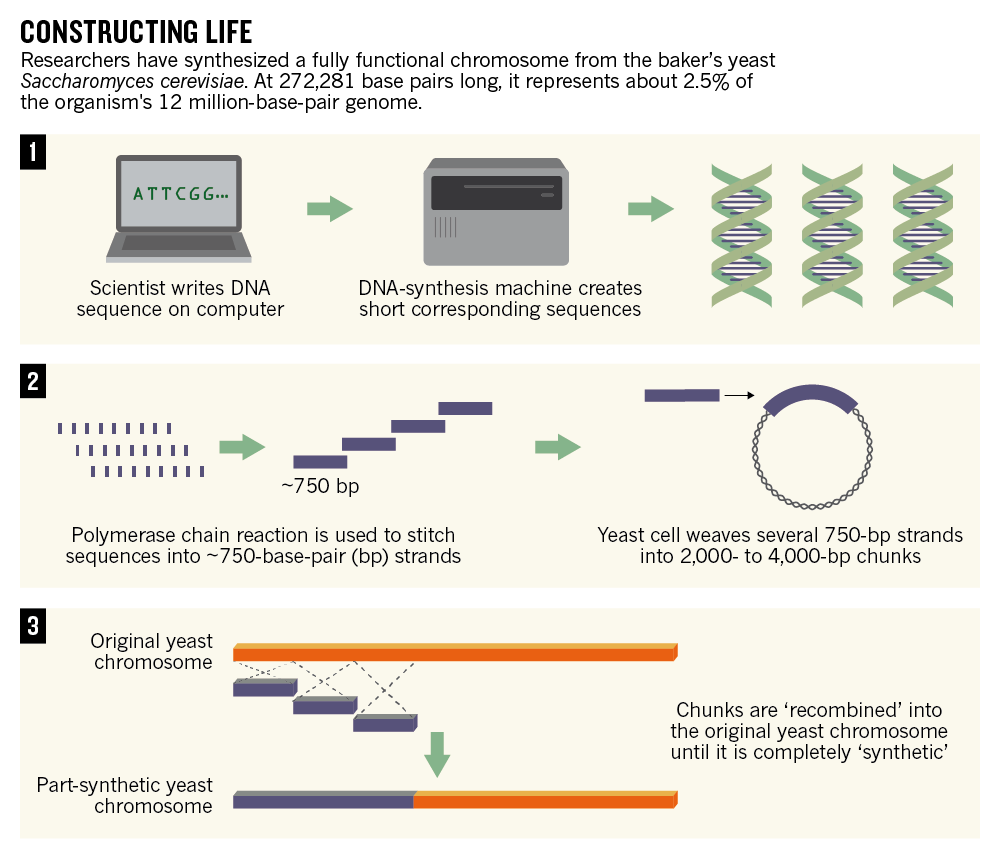
Designing the first fully synthetic yeast chromosome. Nature News
How do scientists program cells to act like a computer?
This is an approach that has garnered a lot of interest. It's essentially designing genes to function in logical circuits, sort of like computers. It has attracted work from many academic groups and startups. And even high school students are now participating in the yearly synthetic bio iGEM competition, which included 245 teams in 2013.
Here's the idea. Genes can be thought of like an input/output system that already does some simple logic. The inputs are molecules that interact with genes to help turn them on or off. The outputs are what the gene makes after it's turned on — usually a protein of some sort. For example, the gene for the enzyme that digests lactose naturally turns on whenever there's lactose around, but not glucose.
Scientists have come up with clever ways to manipulate, combine, and tweak these stretches of DNA to do some pretty interesting things.
In 2012, Swiss researchers showed that they could get mammalian cells to do math. They created genes that only turn on if two particular inputs are there at the same time — so that the genes essentially compute an "AND" function. And they made others that compute other functions. By combining basic logical functions — "AND," "OR," "NOT," and combinations of them — they got cells to do binary addition and subtraction like computers do and then show the right answer by glowing red or green. Others have done projects that also involved memory.
In another example (pictured), a team from the University of California at San Francisco created a plate ofE. coli bacteria that can sense and then trace out an edge of a picture. It's a demonstration of simple logic that could someday get built up into far more complex code. The logic they programmed is as follows: (1) If you sense light, make a certain cell signaling molecule. (2) If you're sensing the signaling molecule (meaning you're near a cell that's in the light) and are not yourself sensing light, then manufacture a dark pigment.

These E. coli have been engineered so that they can find and trace an edge by producing a dark pigment. Popular Science
Researchers have also made DNA elements that are toggle switches that can be turned on or off, ones that reduce noise in response to negative feedback, and ones that create an oscillating signal, among others.
There are now thousands of such interchangeable building blocks held in various databases, such as the public one run by the BioBricks Foundation. The idea is to use these tools to engineer living machines that can perform a variety of tasks.
What about changing the molecules of DNA itself?

Researchers are changing the molecules that make up DNA. UIG via Getty Images
Normally, the cellular factories that construct proteins from DNA instructions are doing so from a limited number of types of parts. There are only 20 standard amino acids — the building blocks that make up the estimated 19,629 human proteins.
But what if an engineer wants to use a lab-made amino acid, a new widget that's never been seen in nature?
First, they'd have to mess with DNA. The DNA that codes for proteins is read three letters at a time, and all of DNA's four letters (A, C, T, G) already have a hard translation for what amino acids they code for. And all of the combinations are already taken.
So, in order to use new amino acids, some engineers want to expand the DNA alphabet with even more letters. This is tricky because it requires retrofitting artificial DNA letters onto eons-old cellular machinery.
In May, 2014, researchers published in Nature that after screening some 300 possible new DNA letters, they found ones that E. coli bacteria would accept. They called these new letters X and Y. The bacteria were able to use their existing machinery to copy DNA containing X and Y for 24 generations (about 15 hours). But researchers have only shown that the cells could copy the DNA, not actually use it. Next up, they'll need to demonstrate that they can get cells to read these new letters to actually make proteins.
Other groups have been focusing on the chemical backbone that holds DNA together. They've created DNA with several other backbones, called XNAs, and have shown that they can get cells to accept and copy them. One possibility is to use such techniques to make DNA that's hardier and more resistant to degradation.
What are the major challenges in synthetic biology?
1) Designer cells can evolve — in unpredictable ways
As helpful as evolution has been for actual life in the real world, life's ever-changing nature is annoying if you're trying to engineer life to become a predictable tool.
As helpful as evolution has been for actual life in the real world, life's ever-changing nature is annoying if you're trying to engineer life to become a predictable tool.
Here's why: cells acquire random mutations in their DNA. And some cells will produce more offspring than others or completely die off. The result is that every new generation is slightly different than the one before. That can be an annoyance if, say, you are trying to design cells to perform a specific task in a pharmaceutical factory.
2) Cells are very messy
Another challenge is that cells are far more disorganized than a circuit board or computer program. The elements of a circuit board can be lined up in a precise order so that the output of one element can be funneled straight into the input of the next.
Another challenge is that cells are far more disorganized than a circuit board or computer program. The elements of a circuit board can be lined up in a precise order so that the output of one element can be funneled straight into the input of the next.
But a cell is an altogether messier situation. The molecules in a cell, including those that people are using as inputs and outputs, are generally lumped together in the same space and — literally — jiggling around randomly. So there's a way higher chance of something cross-reacting, and that can cause problems.
3) Mammals' cells are difficult
A third challenge is that cells from more complex creatures, like mammals, tend to be far more difficult to engineer than, say, bacteria.
A third challenge is that cells from more complex creatures, like mammals, tend to be far more difficult to engineer than, say, bacteria.
Mammals' cells, for example, usually have two copies of each gene in a cell, whereas bacteria generally have one. Also, the processes that regulate what genes get used are more multilayered and complicated. And inserting and deleting genes in mammals' cells is also far more difficult. (Although in the past few years, a new gene editing system called CRISPR has made deleting genes easier.)
How would I know if my food contains synthetic biology products?
You generally wouldn't know. There's no federal law that requires ingredients from synthetic biology to be labeled. This is the case in the US for all genetically engineered foods, including GMO corn and soy and products made from them.
Several ingredients produced by synthetic organisms (but not actually containing these organisms) are on the market in soaps, cosmetics, and foods. You can read a good review of what's going on with these ingredients and (non-)transparency about them in this New York Times story here.
Isn't there a risk that these artificial cells could escape into the wild?
That's one concern, although researchers aren't usually in the habit of simply sending these organisms to the dump without precautions.
There are generally rules in place for them to kill any lab organisms before disposal, generally in a high-temperature, high-pressure oven called an autoclave. (Even a dead lab mouse that hasn't been genetically altered gets autoclaved first, too.)
In some cases, researchers have made organisms that can only survive in the lab — by, for example, tweaking them to need a nutrient that doesn't exist in the wild. It's also possible that scientists could program a kill switch that would turn on at a certain point. (So, for instance, a cell designed to kill cancer could be programmed to self-destruct after it's done its job.)
Is this going to be one more technology that only the rich will get to use while the rest of the world suffers?
Well, only time will tell. Legally, it's possible to patent most of the things that these people are doing. But it's not necessarily the case that that's how synthetic biology will play out. Many people support an open source model, where all information is free for everyone to use.
In a recent piece in Nature, writer Bryn Nelson describes this debate as a clash between engineers and computer scientists, who tend to favor the open source model, and biotech people, who often argue that patents provide economic incentives for innovation.
So far, synthetic biology has been using both. For example, the people uploading genetic sequences into the BioBricks catalog must affirm that they won't claim the sequences as their own intellectual property. But most companies making commercial products, like drugs and food ingredients, are working under the patent system.
More on synthetic biology:
- Profile of and a Q&A with Craig Venter, the biggest name in synthetic bio
- Profile of Jim Collins, synthetic biology pioneer
- The New Yorker wrote a big piece on synthetic biology in 2009. Old, but still good.
- The debate over unlabeled synthetic biology products on the market
Source: Vox